Identification of Conserved Regions in HIV-1 DNA Mutational Patterns Using Multiple Sequence Alignment
DOI:
https://doi.org/10.5530/ctbp.2025.3s.5Keywords:
HIV-I, Mutational Pattern, Conserved region, Big Data, Multiple Sequence Alignment, Phylogenetic TreeAbstract
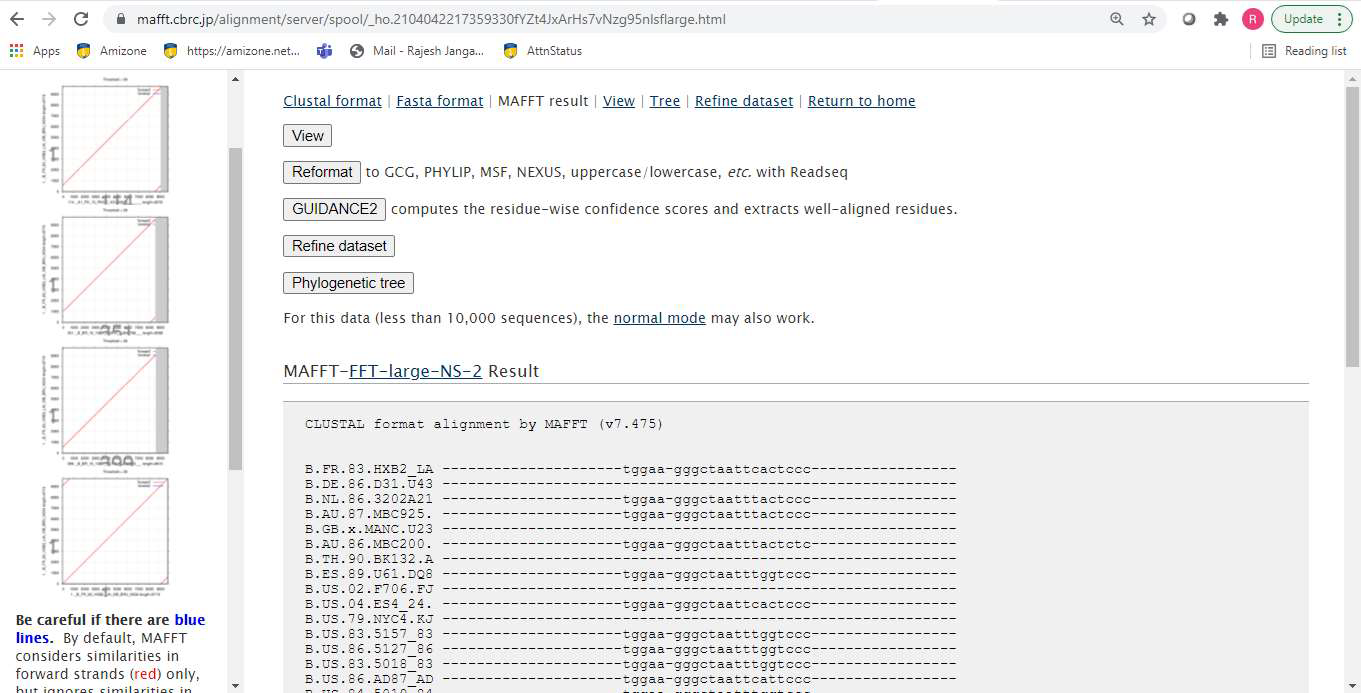
HIV (Human Immunodeficiency Virus) targets and weakens the immune system. If left untreated, it can progress to AIDS (acquired immunodeficiency syndrome), a condition marked by a high viral load and increased infectiousness. Without treatment, individuals with AIDS typically survive about three years. While there is no cure for HIV, it can be managed effectively with proper medical care. This study focuses on identifying conserved regions within the genetic and protein sequences of HIV-1 from large-scale data. By leveraging multiple sequence alignment and machine learning techniques, conserved regions in the mutational patterns of HIV-1 are identified. These conserved regions hold significant potential for the pharmaceutical industry, particularly in development of vaccine.